HSP90: Species Variation
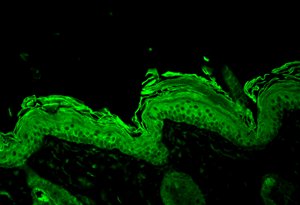
Hsp90 beta visualized on Bouin’s fixed paraffin embedded backskin sections of transgenic mice, using Anti-Hsp90 beta (polyclonal)
The 90 kDa family of molecular chaperones has homologs in eukaryotes like S. cerevisiae, C. elegans, D. melanogaster and many eubacteria, but not in Archaebacteria. The prokaryotic ortholog, HtpG shows 42% identity in amino acid sequence to human HSP90s 14. Although not essential for growth, deletion of HtpG results in slower growth at high temperatures 44 and a slight increase in protein aggregation in heat-stressed cells 45. HtpG is a dimeric phosphoprotein with functional and structural similarities to eukaryotic HSP90s including capability to oligomerize. Unlike eukaryotic Hsp90s, HtpG has a higher thermostability 14, 46. Pro- and eukaryotic HSP90s share the same domain structure: an N-terminal domain (NTD) that binds and hydrolyzes ATP, a middle domain (MD), and a C-terminal domain (CTD) that is important for dimerization 22. In contrast to eukaryotic HSP90s, HtpG lacks the charged linker that connects the NTD with the MD and a C-terminal extension of approximately 35 amino acids. This C-terminal extension contains the conserved M-E-E-V-D amino acid motif involved in the binding of several co-chaperones 47, 48. The crystal structures of the Hsp90/co-chaperone complex from HtpG 21, full-length yeast Hsp90 in complex with ATP 22, Hsp90 complexed to the client proteins Cdc-37 and Cdk-4 49 as well as an mammalian Grp94/nucleotide complex 50 revealed dramatic conformational changes of the chaperones accompanying the ATPase cycle. The crystallographic conformations have been found to vary between a closed ATP-bound form in yeast 22, a V-shaped apo-form and a semi-closed ADP-bound form in HtpG 21, as well as an intertwined closed conformation in Grp94 50. Structural and biochemical studies clearly demonstrated that apo-HtpG may fluctuate between open and extended states while preserving the constitutive CTD dimerization 51, 52. It is interesting to note that HtpG forms highly compact dimers in the presence of ADP although apo-HtpG favors the open conformational state 21. Even in the presence of the ATP analog AMP-PNP, HtpG is capable of maintaining an equilibrium between open and closed states 21. Recent investigations in bacteria, yeast and humans provided evidence for a co-existence of the open and closed conformations during a dynamic equilibrium in the apo, ADP, and AMP-PNP states across diverse species 53, 54.
HSP90 proteins have been identified in many plant species including Arabidopsis thaliana, maize, tomato, orchardgrass, rice, and rape 55, 56, 57, 58, 59, 60. As summarized by Xu and colleagues, plant HSP90s play important roles in plant development, environmental stress response, as well as disease and pest resistence 61. In A. thaliana, seven members of the HSP90 family located to different subcellular compartments have been identified by genome sequence analysis 3. Amongst them, four are found in the cytoplasm and nucleus (AtHsp90-1 to 90-4), and one in mitochondria (AtHsp90-6), ER (AtHsp90-7), and chloroplasts (AtHsp90-5) 3. AtHsp90-2 to -4 have high similarity with homology of approximately 96% suggesting functional redundancy 55, 62. In contrast, sequence identities between cytosolic and other subcellular localized AtHSP90s are at an average of 50% 3. Nevertheless, the NTD as well as the MD have been found as being highly conserved in any AtHsp90 isoform 3. The charged linker region connecting NTD and MD is present in all AtHSP90s except AtHsp90-5 and -6, although both molecules harbour the other signature sequences 3. While AtHsp90-1 to -4 contain the C-terminal target signal M-E-E-V-D for cytosolic location, AtHsp90-5 to -7 contain the N-terminal signal peptide necessary for transport into certain organelles 55, 62. Among the AtHSP90s, AtHsp90-1 is stress-inducible and shares comparatively low sequence identity with the constitutively expressed AtHsp90-2 to -4 63, 64. A comprehensive function/structure analysis of AtHSp90-1 and AtHSp90-3 revealed that AtHsp90-1 exists in several oligomeric states whereas AtHsp90-3 exist predominantly in a high oligomeric state 64. High oligomeric AtHsp90-1 was found to elicit higher holdase chaperone activity than the corresponding monomeric or dimeric states. Furthermore, AtHSp90-3 displayed a higher foldase activity with concomitant high ATPase activity compared to AtHsp90-1. From these findings the authors conclude that, depending on structural discrepancies, cytosolic AtHsp90-1 and -3 elicit different functions suggesting functional divergence regardless their identical subcellular location 64.
In the yeast S. cerevisiae, two genes, HSC82 and HSP82, encode HSP90 proteins that show an identity of 97% 30 but a differential regulation 65. Under physiological conditions Hsc82 is present in 20-fold excess over Hsp82 whose expression is induced more than 20-fold in response to heat shock 66. Both chaperones play an eminent role in yeast translational control and necrotic cell death 67, 68, 69, 70. The group of Paula Ludovico unveiled the relevance of yeast HSP90 isoform translation during apoptosis implying opposing roles for the HSP90 isoforms in cell survival and death 71. While Hsc82 obviously promotes survival, Hsp82 functions as a pro-apoptotic molecule 71.
C. albicans expresses a single heat-inducible HSP90 gene encoding an Hsp90 homolog differing by two amino acids from S. cerevisiae’s HSP90s. The HSP90 gene was originally characterized by the group of Alistair Brown as being essential for viability 31. Later work by the Cowen laboratory established that Hsp90 enables the emergence of drug resistance by stabilizing the protein phosphatase calcineurin and the mitogen activated protein (MAP) kinase Mkc-1 72, 73, 74. Additionally, Hsp90 has been found to regulate biofilm dispersion and drug resistance and to be required for virulence 75, 76. Due to the crucial role of Hsp90 in the C. albicans chaperone network, targeting Hsp90 has been proposed as an effective therapeutic strategy in fungal diseases 77.
Higher eukaryotes such as zebrafish (Danio rerio) express three HSP90 genes. Genome sequence analysis revealed that zebrafish contains two HSP90A genes, hsp90a1 and hsp90a2, closely linked on the same region of chromosome 20, separated by only 698 base pairs from the polyadenylation sequence of Hsp90α1 to the transcription start site of Hsp90α2 29. Hsp90α1 encodes a protein of 725 amino acids, whereas Hsp90α2 encodes a protein of 734 amino acids. Sequence comparison revealed that Hsp90α1 and Hsp90α2 share 88% identity at the mRNA level and 91% identity at the protein level. The major difference between Hsp90α1 and Hsp90α2 lies between positions 237-277 that contain many charged glutamic acid and lysine amino acid residues. Sequence alignment revealed that Hsp90α2 is more closely related to human and mouse Hsp90 and thus most probably represents the Hsp90α ortholog in zebrafish. Du et al. could also demonstrate that Hsp90α1 represents a more diverse homolog of Hsp90α in zebrafish 29.
It has been demonstrated previously that Hsp90α1 is specifically expressed in developing somites and skeletal muscles of zebrafish embryos 78. The group of Yongwang Zhong provide further evidence for the essential role of Hsp90α1 in myofibril organization in skeletal muscles of zebrafish embryos 29. Knockdown of Hsp90α1 resulted in paralyzed zebrafish embryos with poorly organized myofibrils in skeletal muscles, whereas depletion of Hsp90α2 did not affect myofibrillogenesis 29. In previous experiments, knockdown of Hsp90α1 led to significant myosin degradation and up-regulation of the myosin chaperone Unc-45b that is specifically expressed in developing somites and skeletal muscles of zebrafish embryos 79. As demonstrated by Etard and co-workers, knockdown or mutation of Unc-45b resulted in paralyzed zebrafish embryos with muscle defects in thick filaments in the same manner as the lack of Hsp90α1 81. Together, these results underline the crucial impact of Hsp90α1 in muscle development of zebrafish by facilitating myosin folding and assembly into organized myofibril filaments.
Unlike Hsp90α1, Hsp90β is not required for myofibril formation in skeletal muscles 516. It is expressed predominantly in the developing brain, tail bud and cells surrounding the posterior margin of the yolk tube and to a minor degree in the musculature as well as in the embryo80, 81. During early embryogenesis, the expression of the hsp90a gene is strongly up-regulated during heat shock in zebrafish whereas expression of the hsp90b gene appears to be weakly induced 28.
In mammals as well as in zebrafish three isoforms of cytosolic HSP90s are present, while C. elegans and Drosophila contain only one Hsp90 homolog (Table 2). The Drosophila Hsp90, Hsp83 was initially discovered during work on the salivary glands of the fruit fly whose expression is up-regulated upon heat stress 82, 83, 84. Hsp83 interacts dynamically with various co-chaperones modulating its substrate recognition, ATPase cycle and chaperone function. Together with Hop and piwi, Hsp83 mediates canalization, also known as developmental robustness, most likely due to epigenetic silencing of existing genetic variants and suppression of transposon-induced new genetic variation 85, 86. In contrast to other major heat shock proteins, Hsp83 from D. melanogaster is also expressed at normal growth temperatures and is also developmentally expressed during oogenesis. Apart from its chaperoning function, Hsp83 has been found to function as a regulator of the heat shock factor HSF-1, which is the main activator of the stress response 87, 88, 89. It is interesting to note that in Drosophila cells, the reduction of Hsp83 levels led to the induction of the heat shock response 90. Hsp90 can therefore be considered as being a repressor of the heat shock response in Drosophila.
The genome of the unicellular green alga Chlamydomonas reinhardtii encodes three Hsp90 chaperones, termed CrHsp90A to -C 91. CrHsp90A and CrHsp90B have been found to localize to the cytosol and to the ER, respectively, while CrHsp90C may be targeted to the chloroplast and/or to mitochondria 91. A more recent approach revealed that CrHsp90C is targeted only to the chloroplast 92. CrHsp90C is expressed under basal conditions and is strongly induced by heat shock and moderately by light. Furthermore, CrHsp90C exists mainly as a dimer in cellular extracts, but also forms high molecular mass complexes with several other proteins such as the plastidic CrHsp70B chaperone 92. CrHsp90C and the higher plant plastidic HSP90s contain a D-P-W motif at their C-terminus, which is absent in mitochondrial Trap-1 2 and in HSP90s localized to ER and cytosol 92. The charged linker connecting NTD and MD of Hsp90 is present in all Chlamydomonas and higher plant plastidic HSP90s, although being shorter in the latter 3, 92. Less information is available for the ER-specific CrHsp90B which harbours the ER-targeting motif K-D-E-L at the C-terminus 91.
Phylogenetic analyses imply that organellar HSP90 genes of modern eukaryotes originate from duplications of ancient HSP90 genes, that acquired sequences required for organellar targeting of peptides in a secondary event, whereas the endosymbionts’ htpG genes have been lost 36, 93. As described elsewhere, gene duplications giving rise to mitochondrial HSP90 genes were obviously manifested only in some lineages, thereby explaining the presence of genes encoding mitochondrial HSP90s in a couple of organisms such as A. thaliana, D. discoideum, Drosophila spec., and humans 2, 3, 94, and the absence in others, like C. thaliana and Saccharomyces spec.