HSP90: Protein Type
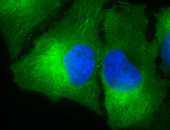
 Hsp90 (total) visualized on a human keratinocyte line HaCat, using Anti-Hsp90 (total) (clone: 4F3.E8)
Hsp90 (total) visualized on a human keratinocyte line HaCat, using Anti-Hsp90 (total) (clone: 4F3.E8)
The HSP90 family represents a group of well-conserved proteins with an average molecular mass of 90 kDa. There are two major cytosolic Hsp90 isoforms, the inducible Hsp90α1 (HspC1) and the constitutive Hsp90β (HspC3), commonly termed Hsp90 1,26. Hsp90 is the major soluble protein of the cell and most commonly located in the cytoplasm. Like many other chaperones, Hsp90 is a rather hydrophobic protein whose hydrophobicity further increases after heat shock 32,33. The HSP90 family members are encoded by a multigene family encompassing six genes and 11 pseudogenes in humans 26. Functional genes encoding HSP90 proteins map to human chromosomes 6, 11, 12, 14, and 16 26. The most studied genes are HSP90AA1 (HSPC1) and HSP90AB1 (HSPC3) encoding proteins that differ by eight amino acids. While Hsp90α1 (HspC1) represents the stress-inducible isoform, Hsp90β (HspC3) is a constitutively expressed protein. The genes are clustered on chromosome 14q32.32 and 6p12, respectively. Two transcript variants encoding different isoforms have been found for the HSP90A gene.
Grp94 (Hsp90B1, HspC4) is the longest member of the HSP90 family with 803 amino acids characterized by three unique deletions and three unique insertions showing approximately 42% identity and 60% similarity with Hsp90α1 and Hsp90β, respectively 26. It harbours the highly conserved C-terminal sequence K-D-E-L facilitating its retention in the ER 34.
Trap-1 (Hsp90L, HspC5) is the shortest family member containing five unique deletions and four unique insertion sites but lacking the charged conserved middle domain. Trap-1 has an average 43% identity and 60% similarity with Hsp90α1 and Grp94, respectively. Functionally important residues such as Lys231 and Phe263 are absent in Trap-1 while Ser231 and Ser263 are present in Hsp90α1 and Hsp90β only. Members of the HSP90AA and HSP90AB subclasses do not harbour a signal peptide although 21 amino acid residues of the Grp94 signal peptide and approximately 30 amino acid residues of the Trap-1 signal peptide have been predicted 26. The molecular masses among the HSP90 family members range from ≈ 12 – 98 kDa in the HSP90AA subclass, ≈ 8 – 85 kDa in the HSP90AB subclass, ≈ 33 – 90 kDa in the HSP90B (GRP94) subclass, and ≈ 54 – 77 kDa in the TRAP subclass. As predicted by the work of Chen and colleagues, various conservative and/or functional regions may be involved in mediating Hsp90-specific functions. These regions comprise an amidation site, a leucine zipper domain, an N-myristoylation site, an N-glycosylation site, a tyrosine sulfation and a tyrosine phosphorylation site as well as phosphorylation sites for cAMP and cGMP-dependent protein kinases and casein kinase II. In most of the HSP90 family members examined, distinct small “signature” sequences can be found including an ATP-binding domain as well as a glutamic acid-rich motif. Unlike the TRAP subclass members, a four-helical cytokine domain can be found in the HSP90AA, HSP90AB, and HSP90B subclasses while the ER-targeting motif K-D-E-L is present in Grp94 (HspC4, Hsp90B1).