HSP90: Figures
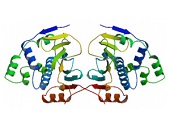
Figure 1. Structure of the tetragonal form of the N-terminal domain of Hsp90 from yeast (PDB ID: 1ah6).
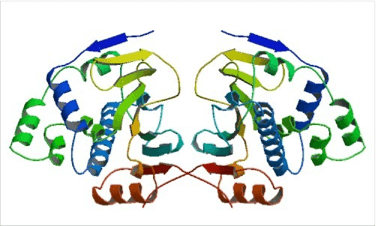
Figure 1. Structure of the tetragonal form of the N-terminal domain of Hsp90 from yeast (PDB ID: 1ah6).
The conformation of the N-terminal domain (NTD) of the yeast Hsp90 shows a dimeric structure based on a highly twisted sixteen stranded beta-sheet, whose topology suggests a possible 30-domain-swapped structure for the intact Hsp90 dimer. The opposing faces of the beta-sheets in the dimer define a potential peptide-binding cleft, indicating that the NTD serves as a molecular ‘clamp’ in the binding of ligand proteins to Hsp90 514. Critical amino acids within NTD directly involved in interaction of yeast Hsp90 with ATP are Asn37, Asp79, Lys98, and Phe124. The ATP-binding domain within the NTD also functions as interation site of diverse natural substances such as the antibiotics radicicol and geldanamycin that promote degradation of protein kinases and interfere with Hsp90 functions.
Figure 2. Structure of full-length yeast Hsp90.
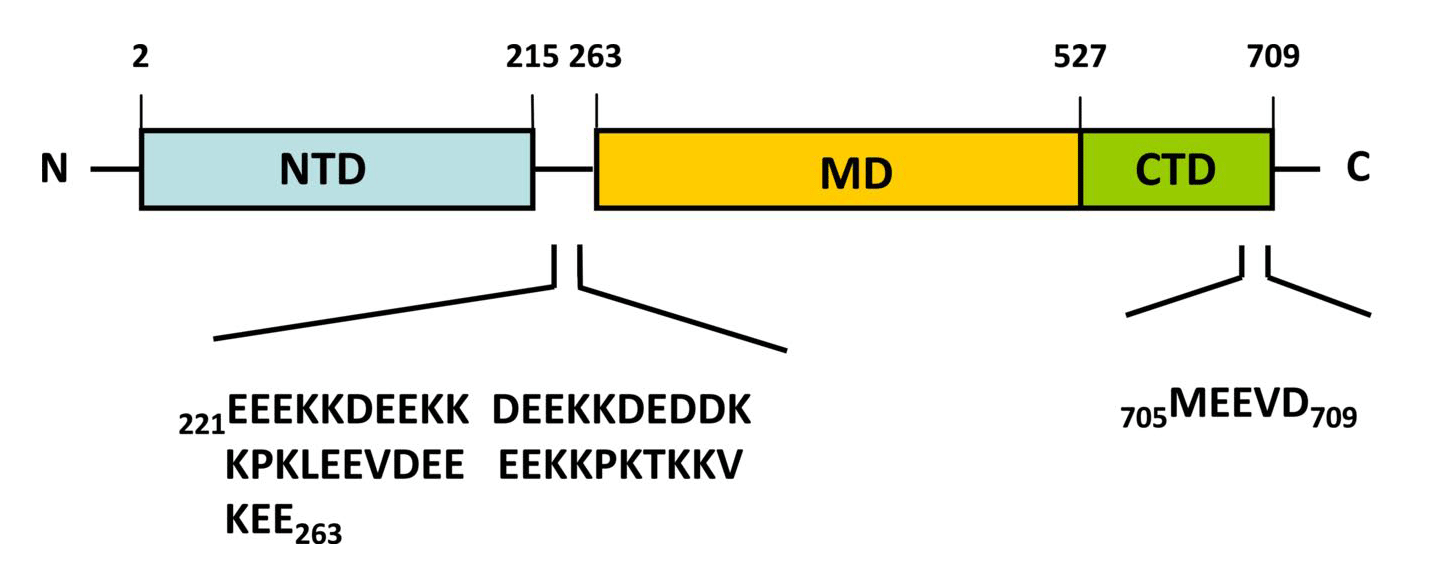
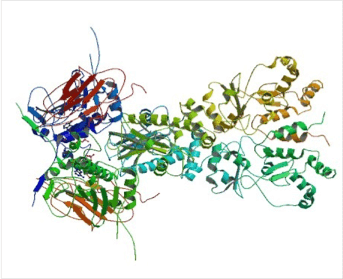
Figure 2. Structure of full-length yeast Hsp90.
(Top) Schematic representation of the yeast heat-inducible Hsp82. Each monomer is formed by an N-terminal nucleotide binding domain (NTD) with ATPase ativity (residues 2-215), a middle domain (MD; residues 262-526) and a C-terminal dimerization domain (CTD; residues 527-709) harbouring the TPR repeat-binding motif M-E-E-V-D. NTD and MD are linked by a short highly charged region of 5 x 5 amino acid repeats consisting of [DE]-[DE]-[DE]-K-K. (Bottom) Crystal structure of dimeric full-length yeast Hsp90 in complex with an ATP analogue and the co-chaperone p23/Sba-1 (PDB ID: 2CG9). The structure reveals the complex architecture of the ‘closed’ state of the Hsp90 chaperone 22.
Figure 3. Structures of the full-length Hsp90 dimer.
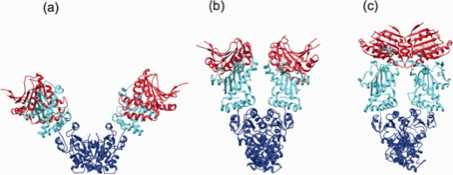
Figure 3. Structures of the full-length Hsp90 dimer.
Ribbon and tube representation of the open (a), partially closed (b) and closed (c) conformation of Hsp90 dimers. The models are based on crystal structures of full-length E. coli Hsp90 HtpG (PDB ID: 2IOQ 21), full-length mammalian Grp94 in complex with the ATP analog AMP-PNP (PDB ID: 2O1U 50) as well as full-length yeast Hsp90 in complex with ATP (PDB ID: 2CG9 22). The N-terminal nucleotide-binding domain (NBD) is given in red, the middle domain (MD) in cyan, and the C-terminal dimerization domain (CTD) in blue while bound nucleotides (Figs. b + c) are given in grey (reproduced from Clare and Saibil 515).
Figure 4. Cartoon illustrating the ATPase-driven steroid hormone receptor activation cycle.
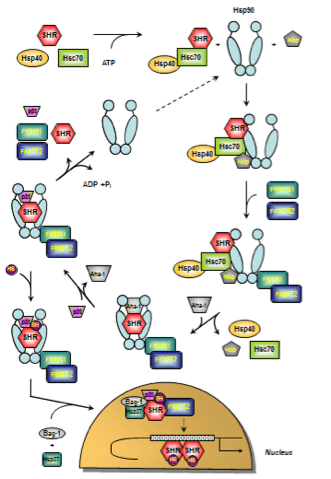
Figure 4. Cartoon illustrating the ATPase-driven steroid hormone receptor activation cycle.
The early stage of foldosome assembly comprises recruitment of the Hsc70/Hsp40 chaperone complex to the steroid hormone receptor (SHR) in an ATP-dependent manner. After docking of the adapter proteins Hsp90 and Hop (Hsp90/Hsp70 organizing protein) to the open-state Hsp90, the intermediate foldosome complex is formed. Maturation of the foldosome complex is achieved by the ATP-dependent addition of FKBP51 and FKBP52 followed by N-terminal dimerization of Hsp90 and loss of Hsc70/Hsp40 and Hop. In this conformation, Hsp90 shows high affinity for p23 whose binding is facilitated by the Hsp90 ATPase activator Aha-1. The SHR is now able to bind the ligand leading to conformational re-arrangements culminating in activation and release of SHR subsequenty followed by nuclear translocation, receptor dimerization and binding to response elements in regulatory regions of certain target genes. Apart from p23 and FKBP51/52, further chaperones and co-chaperones have been described to have nuclear effects on SHR action such as Bag-1, Hsp10, Hsp27, and Hsp60 (figure adapted from Cano et al. 2013 163 and Eckl and Richter 2013 281; for details see text).