HSP90: History
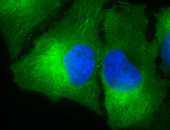
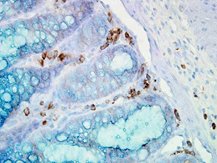 IHC staining of inflammatory cells in mouse colon tissue, using Anti-Hsp90 alpha (clone: Hyb-K41009)
IHC staining of inflammatory cells in mouse colon tissue, using Anti-Hsp90 alpha (clone: Hyb-K41009)
Heat shock protein 90 (Hsp90) was originally described amongst a defined set of heat shock proteins (HSPs) that are rapidly induced in fungal, plant and animal cells in response to acute thermal up-regulation 9,10,11,12. This HSP induction is referred to as the heat shock response (HSR) that is ubiquitous across the bacterial, archaeal and eukaryotic kingdoms 13. In 1987, Bardwell and Craig isolated a gene from E. coli called htpG which is homologous to the gene encoding the Drosophila 83 kDa heat shock protein 2 14. The isolation of an E. coli homolog of Hsp83 illustrates the remarkable conservation of HSPs in evolution. Notably, heat shock-induced htpG gene expression was extensively investigated in continuous cultures of E. coli by Heitzer and colleagues in the early 1990s 15. A full-length cDNA encoding a HSP (AtHsp83) belonging to the HSP90/HSPC family of Arabidopsis thaliana has been isolated and sequenced at the time 16. In this study, the low level of AtHsp83 transcripts in Arabidopsis plants was found to increase rapidly after exposure to elevated temperatures.
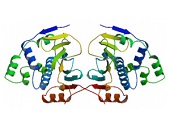
HSPs are subgrouped into families based on their molecular mass. Hsp90 as a member of the HSP90/HSPC family was initially identified as a molecular chaperone suppressing the formation of protein aggregates by binding to the target proteins in an ATP-dependent manner 17. Hsp90 is a ubiquitous and abundant cytosolic chaperone which was further characterized as being critically involved in the function and regulation of steroid hormone receptors 18. However, an extracellular location of Hsp90 was firstly implicated in 1986 when a tumour-specific transplantation antigen from the cell surface of mice tumours was identified as being identical to yeast Hsp82 and Drosophila Hsp83, respectively 19. In 1997, crystal structures of complexes between the N-terminal domain of the yeast Hsp90 chaperone and ADP/ATP led to the identification of a specific adenine nucleotide-binding site homologous to the ATP-binding site of DNA gyrase B 20. The structure of the yeast Hsp90 bound to the ATP analogue AMP-PNP and to the co-chaperone p23 provided insight into the ATP-bound state, while crystallographic studies of the E. coli Hsp90 HtpG made a picture of the nucleotide-free and the ADP-bound state 21, 22. Based on these findings it became obvious that Hsp90 can adopt different conformations triggered by nucleotides. Recent single molecule studies indicated that dimers of Hsp90 perform large conformational rearrangements suggesting the existence of anti-correlated C- and N-terminal dimerizations 23. By using nuclear magnetic resonance (NMR) spectroscopy, the group of Stefan Rüdiger identified binding sites of several co-chaperones and substrate proteins in full-length HSP90s thereby increasing our understanding of the structural organization of HSP90s 24.